Abstract
Background: The IGHV mutation status (IGHVms) is used for classifying CLL as mutated or unmutated depending on the identity of the expressed IGHV gene to the germline template with 98% identity used as cutoff. CLL patients with unmutated IGHV have a worse prognosis. WGS and WTS are comprehensive techniques identifying all genetic alterations in a single approach.
Aim: Evaluate the accuracy of WGS and WTS in determining the IGHVms in a clinical setting.
Patients and Methods: The cohort comprised 216 CLL patients. The diagnosis was established following WHO classification. WGS (100x, 2x151bp) and WTS (5x107 reads, 2x101bp) data were generated on NovaSeq instruments. The identification of IGHV rearrangements (IGHVr) using NGS data was performed using IgCaller for WGS data, and MiXCR for WTS data. The tools were used with default settings.
For each sample, we used the IGHVr clonotype supported by the largest number of reads for further analyses. IGHVr calls were considered matching when the same IGHVr was called in routine diagnostics based on Sanger sequencing of PCR fragments. IMGT/V-QUEST was used to obtain the IGHVms for sequences obtained from PCR fragments and MiXCR. IgCaller reports its own IGHVms.
Results: While MiXCR successfully identified an IGHVr for 98% of samples (211/216), IgCaller made an IGHVr call for only 76% of samples (164/216). In four samples, neither tool could identify an IGHVr. In cases where no IGHVr was identified by MiXCR or IgCaller, routine diagnostics found a mutated IGHV in 5/5 cases and 49/52 cases, respectively.
When an IGHVr was detected using NGS data, it was in good agreement with routine data for both tools. MiXCR produced calls concordant with routine data in 92% of samples with mutated IGHV (92/100) and in 98% of samples with unmutated IGHV (109/111). For IgCaller, the percentage of concordant calls was 95% for mutated samples (52/55) and 98% of unmutated samples (106/108).
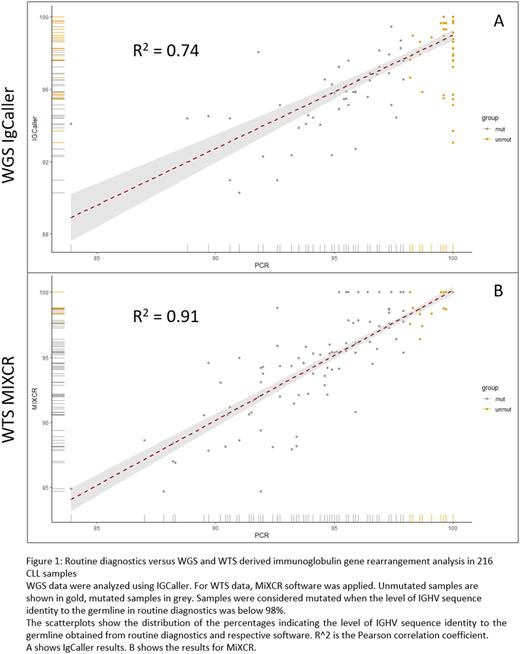
For samples with matching IGHVr calls, we compared the estimates of sequence identity to the germline (Fig.1). The correlation coefficient R2 between the measures provided by IgCaller and routine was 0.74 while the corresponding coefficient for MiXCR was 0.91.
Failure to identify an IGHVr was dependent on the IGHVms. The mean percentage of sequence identity to the germline in samples without an IGHVr call made by IgCaller was 92.9% compared to 98.2% in samples where IGHVr calling succeeded (p=3x10-24). The corresponding numbers for MiXCR were 97.0% in samples with IGHVr call and 93.3% in samples without IGHVr call (p=0.03).
MiXCR was able to successfully identify IGHVr also in samples with high mutation burden, in which IgCaller showed limitations. IgCaller works with WGS data that, in the present study, have been sequenced at a coverage of 100x. MiXCR utilizes RNASeq data and the number of reads covering the rearranged IGHV genes is one to two orders of magnitude higher. Indeed, down sampling the number of reads utilized by MiXCR to 10% and 1% of all reads reduced the number of samples with IGHVr calls to 189/216 and 132/216, respectively.
Taken together, the number of samples where both IGHVms and IGHVr calls were concordant with routine results was 129/216 for IgCaller and 180/216 for MiXCR. In 112/216 samples, all three methods yielded identical results. We found 7 samples with concordant IgCaller and MiXCR calls that nevertheless differed from routine results. These samples showed germline identities close to the 98% cut-off, which led to discordant IGHVms calls between NGS and routine. This observation holds for both tools individually: When the IGHVr call is concordant with routine, discordant IGHVms calls indicate a weakly mutated IGHV.
Conclusions: Detection of IGHVms and IGHVr using NGS data is feasible but cannot guarantee results identical to routine diagnostics yet, particularly in CLL patients with mutated IGHV. WTS produced better results than WGS. Down sampling of WTS data to a level of sequence coverage at IGHV loci comparable to WGS data eliminated this difference. High quality NGS data are critical. For weakly mutated IGHV, discordant IGHVms calls are common. Thus, using either tool alone or in combination cannot be recommended for routine diagnostics use.
Disclosures
Mueller:MLL Munich Leukemia Laboratory: Current Employment. Walter:MLL Munich Leukemia Laboratory: Current Employment. Hutter:MLL Munich Leukemia Laboratory: Current Employment. Nadarajah:MLL Munich Leukemia Laboratory: Current Employment. Dicker:MLL Munich Leukemia Laboratory: Current Employment. Meggendorfer:MLL Munich Leukemia Laboratory: Current Employment. Kern:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership. Haferlach:Munich Leukemia Laboratory: Current Employment, Other: Part ownership. Haferlach:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal